Google – Real-time sepsis diagnostic
Summary
Digging deeper into the smallest details always brings valuable insights. The same is relevant for Genomics, an interdisciplinary field of biology that focuses on the structuring, functioning, evaluating, mapping, and editing of genomes. A genome is an organism’s complete set of DNA, including all of its genes.
The main challenge in modern Genomics is real-time Nanopore DNA sequencer Processing. Despite the availability of nanopore DNA sequence devices, which you can buy for around one thousand dollars, there was no relevant software that could analyze DNA samples in real-time.
At the same time, existing genomic analyses require DNA sample transportation to a centralized facility, sequencing, and analyzing samples in a batch process, which takes weeks and even months. Such a slow analysis process may cost lives, especially for patients with sepsis, for whom each hour of delay decreases chances to survive by 4%, and death can occur in 24-48 hours.
The backstory
Digging deeper into the smallest details always brings valuable insights. The same is relevant for Genomics, an interdisciplinary field of biology that focuses on the structuring, functioning, evaluating, mapping, and editing of genomes. A genome is an organism’s complete set of DNA, including all of its genes.
The main challenge in modern Genomics is real-time Nanopore DNA sequencer Processing. Despite the availability of nanopore DNA sequence devices, which you can buy for around one thousand dollars, there was no relevant software that could analyze DNA samples in real-time.
At the same time, existing genomic analyses require DNA sample transportation to a centralized facility, sequencing, and analyzing samples in a batch process, which takes weeks and even months. Such a slow analysis process may cost lives, especially for patients with sepsis, for whom each hour of delay decreases chances to survive by 4%, and death can occur in 24-48 hours.
It started out when Allen Day, a data scientist and senior developer advocate from Google Cloud Platform, was looking for a cloud architect with a GCP certificate. During the search, Allen got in touch with us. After he ensured that we had a certified GCP architect, (Allen has found only two developers with relevant experience), we started to clarify more of the project’s details.
Allen needed such a specialist for the Queensland University of Technology project. The project should combine Machine Learning capabilities, Google Cloud Platform tools for data analytics, and existing genomic datasets with streaming data analytics.
Initially, Allen needed just one professional from our team. But, with time, Allen appreciated our technological expertise, proactivity, and product mindset, which allowed us to extend the development team to 10 members.
What is real-time DNA sequence analysis application
The application analyses DNA nanopores in real-time. It detects taxonomical proportions, potential viruses, and pathologies, antibiotic resistant genes, etc. Then, the app visualizes the results received via the interactive Sunburst tool. Thanks to convenient and detailed data visualization, medical experts can make an informed decision on a patient treatment plan.
Nanopore DNA sequencing has numerous applications in the Healthcare and Agricultural industries:
- Infection diagnosis from a blood sample and taxonomical counting
- Monitoring, profiling, and tracking the evolution of antibiotic resistance genes
- Pathogen detection in sewage or public health
- Identification of viruses in cassava crops
How does it work
Blood and other DNA samples are collected from a portable nanopore sequencing device. We used a MinION device, developed by Oxford Nanopore Technologies. Then, the received data is sent to the platform and goes through the following workflow:
- Ingestion – files are uploaded to the Google Cloud Platform and streamed into the processing pipeline
- Base-calling stage – machine learning model infers DNA sequences from electrical signals
- Alignment stage – via a DNA database, the samples are analyzed to find pathogen sequences, taxonomy enrichment, and other anomalies
- Summarization stage – calculation of each pathogen’s percentage in the particular sample
- Storage and visualization – the results are saved to Google Firestore DB and subsequently visualized in real-time with D3.js.
Project objectives
To develop an application for DNA nanopore sequencer analysis, we needed to go through the following steps.
- Investigate Genomics domain data formats
- Develop of a raw sequencer data simulator
- Empower the app with a source data reading
- Develop logic for the alignment stage
- Develop flow for the summarization stage
- Find relevant solutions and tools for analyzed data storage and visualization
- Write documentation on how to use the application and make it available on GitHub
Our Challenges
During the development of the DNA nanopore sequencer application, we faced the following challenges:
- Build scalable and reliable system architecture. Nanopore Sequencer DNA Analysis is a resource-demanding procedure. Due to the high volume of data and tight time constraints, the system needs to scale accordingly. We eliminated this challenge by leveraging Google Cloud Platform compute, storage, and data processing tools. Thanks to such an approach, we achieved a smooth, streamlined, and reliable scalability for data processing operations.
- Data processing logic. The project required fine-tuned data processing operation for providing a broad scope of results in minimal time. Thus, we needed to develop a data processing logic that connects the analytics application to the cloud platform and guarantee an effective information turnover. To achieve this goal, we used the Apache Beam library that runs on Google Cloud Dataflow and supports integration with other Google Cloud services. We also integrated the Compute Engine to build the auto-scaling Alignment Cluster in the application.
- DNA Analysis Tools integration. DNA Analysis tools for Nanopore sequencers were just desktop utilities, thus we needed to adjust them for cloud platforms. We also needed to integrate the desktop-based DNA analysis tools into a unified, scalable system. We reinterpreted the desktop-based DNA analysis tools for HTTP format and distributed them as web services so they could process large quantities of data in a shorter time span.
How we did it
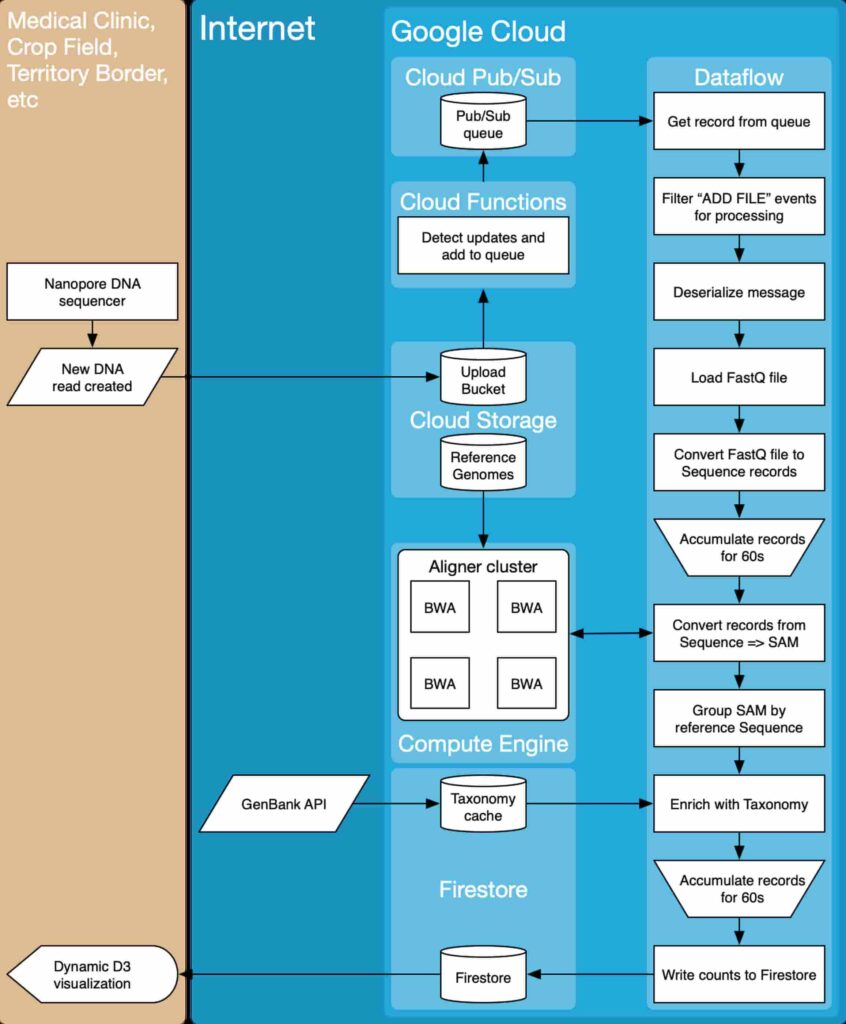
System architecture
For the system architecture, we applied a bunch of Google Cloud Platform data processing tools, as well as compute capabilities. Data collected from a nanopore DNA sequencer becomes available in the New DNA read file and uploaded to a Cloud Storage bucket. Once all files are uploaded, they go through a workflow that converts the input files into actionable reports.
Machine learning engine
The application should include a machine learning algorithm that would analyze genetic engineering databases to come up with relevant DNA test results. For this goal, we applied a machine learning model on TensorFlow, previously trained with several genomic databases.
Server-side
We needed to create a server-side with web and mobile developers in mind. Thus, to store databases and files, we leveraged Firebase, a document-storage system that can represent hierarchical data, essential for representing biological taxonomies.
Client-side
Our primary concern was to shorten the time data is uploaded from the sequencer and visualized.
To keep things as fast as possible from the client-side, we implemented a dynamic dashboard with D3.js, which periodically polls a database for new data and updates the Sunburst chart visualization accordingly.
Our tech stack
Tech Stack:
- JAPSA, a Java Package for Sequence Analysis.
- TensorFlow, an open-source library for training an ML algorithm
- Chiron Base Caller for translating raw nanopore signal
- Samtools utilities for interacting with and post-processing short DNA sequence read alignments in the SAM, BAM and CRAM formats
- BWA MEM algorithm for performing local alignment
- Minimap2, a versatile sequence alignment program for aligning DNA or mRNA sequences against a large reference database.
Google Cloud tools:
- Google Cloud Storage for storing and accessing data
- Google Cloud PubSub for sending and receiving messages between application components at low latency with on-demand scalability
- Google FireStore for storing, syncing, and querying data
- Google Cloud Dataflow to simplify streaming data pipeline development with lower data latency
- Apache Beam for defining and executing data processing workflows
- D3 Data Visualization Library for producing dynamic, interactive data visualizations in web browsers
- JavaScript for building Data Visualisation
Team composition
- 2 Data Engineers
- 1 DevOps Engineer
- 1 Web Developer
Result and prospects
We developed Nanopore DNA sequencers that significantly reduce the time it takes to generate DNA sequence data. Now, medical professionals and genomic engineers can receive sequence data generated from samples (the patient, sewage plant, or crop field) within a few minutes.
The potential for nanopore sequencing is promising. The real-time nanopore sequencing platform we developed has a significant impact on the molecular diagnosis of human disease, in particular, sepsis. Together with the Google Cloud Platform team, we created an open-source set of packages of nanopore sequencers available on GitHub.
This project was just the beginning of fruitful cooperation.
Once, Allen Day visited our office in Kyiv to run a Data Science meetup that was quite successful among data scientists and data engineers.





After a year of working with Allen and the GCP team, our CEO, Pavel Tantsiura visited Google’s office in Singapore to sign an official partnership with Google and The APP Solutions and discuss an upcoming project.

Currently, we are working with Allen on the PopGen project in the Genomics field aimed at processing populations of genomes. The project’s directories with rice and human genome samples are available on the GitHub repository.